Writing the evaluation program in tJava
Procedure
- Double-click tJava to open its Component view.
- Click Sync columns to ensure that tJava retrieves the replicated schema of tClassify.
-
Click the Advanced settings tab to open its view.
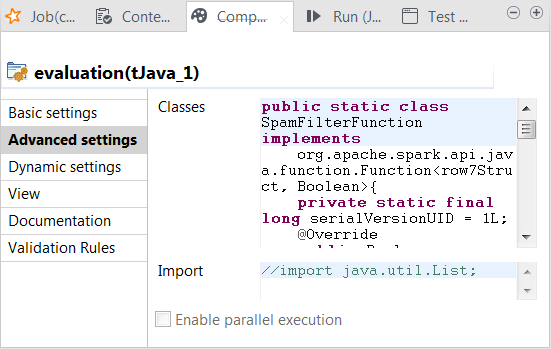
-
In the Classes field, enter code to
define the Java classes to be used to verify whether the predicted class
labels match the actual class labels (spam for junk messages and ham for normal messages). In this scenario, row7 is the ID of the connection between
tClassify and tReplicate and carries the classification result to be sent
to its following components and row7Struct is the Java class of the RDD for the
classification result. In your code, you need to replace row7, whether it is used alone or within
row7Struct, with the corresponding
connection ID used in your Job.
Column names such as reallabel or label were defined in the previous step when configuring different components. If you named them differently, you need to keep them consistent for use in your code.
public static class SpamFilterFunction implements org.apache.spark.api.java.function.Function<row7Struct, Boolean>{ private static final long serialVersionUID = 1L; @Override public Boolean call(row7Struct row7) throws Exception { return row7.reallabel.equals("spam"); } } // 'negative': ham // 'positive': spam // 'false' means the real label & predicted label are different // 'true' means the real label & predicted label are the same public static class TrueNegativeFunction implements org.apache.spark.api.java.function.Function<row7Struct, Boolean>{ private static final long serialVersionUID = 1L; @Override public Boolean call(row7Struct row7) throws Exception { return (row7.label.equals("ham") && row7.reallabel.equals("ham")); } } public static class TruePositiveFunction implements org.apache.spark.api.java.function.Function<row7Struct, Boolean>{ private static final long serialVersionUID = 1L; @Override public Boolean call(row7Struct row7) throws Exception { // true positive cases return (row7.label.equals("spam") && row7.reallabel.equals("spam")); } } public static class FalseNegativeFunction implements org.apache.spark.api.java.function.Function<row7Struct, Boolean>{ private static final long serialVersionUID = 1L; @Override public Boolean call(row7Struct row7) throws Exception { // false positive cases return (row7.label.equals("spam") && row7.reallabel.equals("ham")); } } public static class FalsePositiveFunction implements org.apache.spark.api.java.function.Function<row7Struct, Boolean>{ private static final long serialVersionUID = 1L; @Override public Boolean call(row7Struct row7) throws Exception { // false positive cases return (row7.label.equals("ham") && row7.reallabel.equals("spam")); } } -
Click the Basic settings tab to open its
view and in the Code field, enter the code
to be used to compute the accuracy score and the Matthews Correlation
Coefficient (MCC) of the classification model.
For general explanation about Mathews Correlation Coefficient, see https://en.wikipedia.org/wiki/Matthews_correlation_coefficient from Wikipedia.
long nbTotal = rdd_tJava_1.count(); long nbSpam = rdd_tJava_1.filter(new SpamFilterFunction()).count(); long nbHam = nbTotal - nbSpam; // 'negative': ham // 'positive': spam // 'false' means the real label & predicted label are different // 'true' means the real label & predicted label are the same long tn = rdd_tJava_1.filter(new TrueNegativeFunction()).count(); long tp = rdd_tJava_1.filter(new TruePositiveFunction()).count(); long fn = rdd_tJava_1.filter(new FalseNegativeFunction()).count(); long fp = rdd_tJava_1.filter(new FalsePositiveFunction()).count(); double mmc = (double)(tp*tn -fp*fn) / java.lang.Math.sqrt((double)((tp+fp)*(tp+fn)*(tn+fp)*(tn+fn))); System.out.println("Accuracy:"+((double)(tp+tn)/(double)nbTotal)); System.out.println("Spams caught (SC):"+((double)tp/(double)nbSpam)); System.out.println("Blocked hams (BH):"+((double)fp/(double)nbHam)); System.out.println("Matthews correlation coefficient (MCC):" + mmc);
Did this page help you?
If you find any issues with this page or its content – a typo, a missing step, or a technical error – please let us know!
